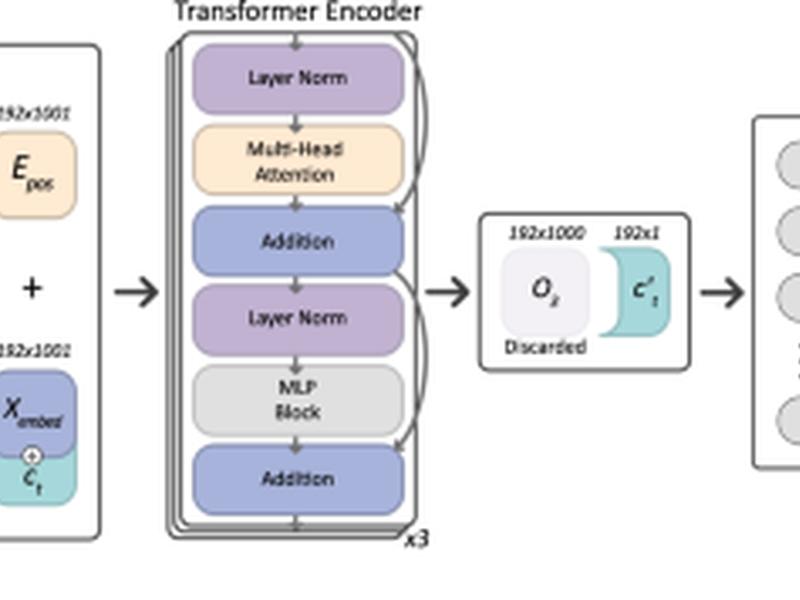
This article presents ECOLE, a deep learning-based method for somatic and germline CNV calling on WES data. ECOLE is based on a variant of the transformer model, which is the state-of-the-art approach to process sequence data in the natural language processing domain. ECOLE improves the exon-wise precision and recall of the next best method’s performance substantially on a benchmark of automated WGS calls. Additionally, ECOLE can be adapted to call somatic variations using transfer learning and fine-tuning the model parameters using a small number of human expert-labeled samples.